AI-Driven Research in the Musser Lab
Research Scientist Kejue Jia is a computational scientist specializing in the application of large language models (LLMs) to address biological problems, with a particular focus on protein language models (pLMs). He leads the development of advanced AI-driven methodologies in our lab, including the pretraining of novel protein and single-cell language models, the fine-tuning of existing foundation models, and the establishment of close computational–experimental collaborations across the group.
Our lab has built robust and mature computational frameworks that integrate AI-based methods into day-to-day biological research, which enables systematic discovery across molecular and cellular scales. The major AI-based approaches currently driving our research include the following:

Kejue Jia (pictured left)
1. pLM-Based Remote Homolog Detection
Identifying homologous proteins in non-model organisms remains challenging, as traditional sequence alignment–based methods often fail at large evolutionary distances. Protein language models, however, capture deep, contextual patterns in protein sequences, enabling the detection of remote homologs with substantially improved sensitivity and precision.

In our lab, we systematically assemble complete animal proteomes and convert protein sequences into optimized pLM embeddings using the Protein Ortholog Search Tool (PROST) workflow (https://github.com/MesihK/prost). This strategy allows us to efficiently identify remote homologous proteins across diverse animal lineages and provides a powerful framework for phylogenetic and evolutionary analyses of gene modules.
2. Deep Learning Model for Protein–Protein Interaction Prediction
For the purpose of efficiently predicting protein interactions, we have developed a novel encoder-only architecture language model trained on experimentally resolved protein complex structures. The model employs a selective masking strategy that forces learning of amino-acid interdependencies specifically at protein–protein interaction interfaces.
The model is subsequently fine-tuned using binary-labeled protein–protein interaction datasets tailored to specific biological case studies. In practice, we integrate whole-body single-cell expression atlases from diverse species, along with newly generated cell atlases from our own lab, to infer protein co-expression patterns. For co-expressed proteins, we apply our model to predict their interactions, which are then assembled into protein interaction networks for specific gene modules comprising large numbers of proteins. These results lead to mechanistic insights into cellular function.

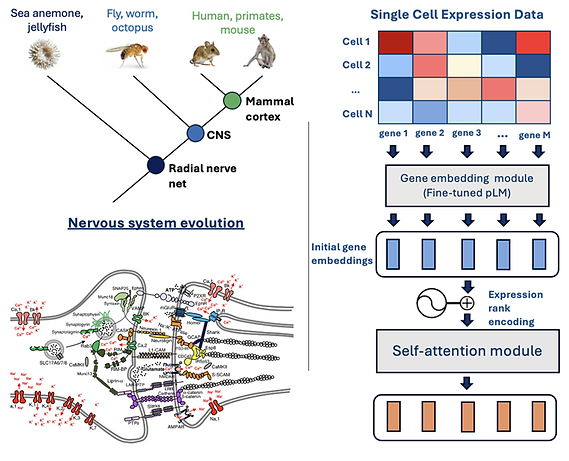
3. Reconstructing the Evolutionary Assembly of Synapses with a Multimodal Language Model
A major pilot initiative in our lab seeks to reconstruct the protein functional modules of the first animal synapse and to elucidate how this architecture was subsequently modified to enable major evolutionary innovations, including the emergence of centralized nervous systems and the mammalian cortex.
To achieve this, we are also developing a novel multimodal language model that integrates large-scale proteomic and single-cell expression datasets from a broad diversity of animals. This approach provides a powerful new lens for uncovering the fundamental design principles of neuronal synapses.
We anticipate that this AI framework will have broad applicability for discovering previously unknown cellular machinery across the tree of life, laying the foundation for an expanded research program that leverages biological diversity to drive advances in cell biology, molecular biology, synthetic biology, and therapeutic discovery.
Finally, these AI-driven frameworks not only accelerate discovery within our own research but also establish a versatile platform for collaborative exploration across disciplines. We actively welcome collaborations with experimentalists, computational scientists, and interdisciplinary teams who share an interest in advancing data-driven biology.
